The definition of “mutation bias”
Mutation bias: a systematic difference in rates of occurrence for different types of mutations, e.g., transition-transversion bias, insertion-deletion bias
Brandolini’s law: it takes 10 times the effort to debunk bullshit as to generate it
If I were to misdefine “negative selection” or “G matrix”, evolutionary biologists would go nuts because theories and results that are familiar would be messed up by a wrong definition. Likewise, a wrong definition of mutation bias is obvious to those of us who are actual experts, because it induces contradictions and errors in things we know and care about.
The actual usage of “mutation bias” by scientists is broadly consistent with a systematic difference in rates of occurrence for different types of mutations and is not consistent with a forward-reverse bias or with heterogeneity in rates of mutation for different loci or sites. To demonstrate this, here is a simple table showing which meanings fit with actual scientific usage, starting with the 3 types of mutation bias invoked most commonly in PubMed (based on my own informal analysis), and continuing with some other examples. The last two refer to the literature of quantitative genetics, which occasionally makes reference to bias in mutational effects on quantitative traits (either on total variability, or on the direction of effects).
| Effect called a “mutation bias” in the literature | Heterogeneity per locus (or site) | Forward-reverse asymmetry | Systematic diff in rates for diff types |
| Transition bias | No | No | Yes |
| GC/AT bias | No* | Yes | Yes |
| Male mutation bias | No | No | Yes |
| pattern in Monroe, et al (2022) | Yes* | No | Yes |
| Insertion or deletion bias | No | Yes | Yes |
| CpG bias | No | Possibly | Yes |
| Diffs in mutational variability of traits | Possibly | No | Yes |
| Asymmetric effect on trait value | No | Possibly | Yes |
How does one concept of “mutation bias” cover such heterogeneity? Every mutation has a “from” and a “to” state, i.e., a source and a destination. A variety of different genetic and phenotypic descriptors can be applied to these “from” and “to” states, which means that we can define many different categories or types of mutations. Different applications of the concept of mutation bias always refer to types whose rates differ predictably, but there are many different ways of defining types, so there are many different possible mutation biases.
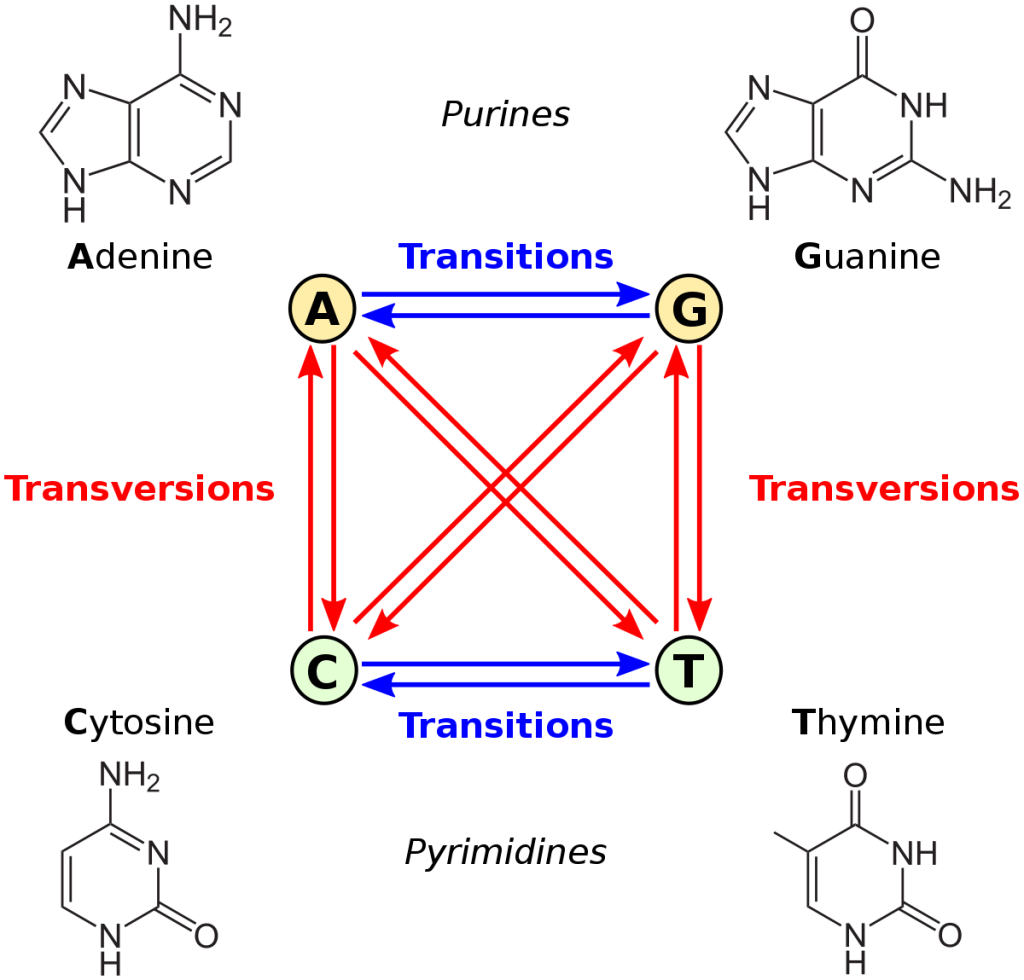
Let’s consider transition-transversion bias, GC vs. AT bias, and male mutation bias. The first is defined relative to the chemical categories of purine (A or G) and pyrimidine (C or T): we apply these categories to the source and destination states, and if they are in the same category, that is a transition, otherwise it is a transversion. The second example, GC/AT bias, is based on whether the shift from the “from” to the “to” increases or decreases GC content. This can be defined either as a forward-reverse asymmetry, or as a difference in mutability of the “from” state, e.g., if A and T are simply more mutable than G and C, the result is a net bias toward GC. In the case of male mutation bias, the categories of mutation are defined by whether the “from” context is male or female.
Note that transition-transversion bias is not a site-wise bias: every nucleotide site is the same in the sense of having 1 transition and 2 transversions (one blue arrow and 2 red arrows in the figure above). Also, transition bias is not a forward-reverse bias, but a difference between two types of fully reversible rates, e.g., under transition bias, the transitions A —> G and G —> A both have a higher rate than the transversions A —> T and T —> A. An insertion-deletion bias is a forward-reverse bias, but it is not a site-wise bias, in the sense that every site has the same set of possible insertions and deletions.
Thus, defining mutation bias as “differences between loci in mutation rates” (Svensson, 2022) is inconsistent with transition bias, GC/AT bias, and male mutation bias, the 3 most familiar and commonly invoked types of mutation bias in the scientific literature. The magnitude of this error is roughly the same as that of defining “genome” as the RNA molecules that store hereditary information. Some genomes are indeed made of RNA. We can imagine a novice RNA virus researcher, e.g., a summer student, who hears everyone in the lab talking about the “genome” which is RNA, and who assumes on this basis that all genomes are RNA, but no experienced scientist who has worked with a variety of organisms or read widely or attempted to teach students would make this kind of error of defining something in a way that excludes the most familiar cases.

Why is this called a “bias”? “Mutation bias” (“mutational bias”, “bias in mutation”) has been a term of art in molecular evolution for over half a century, since Cox and Yanofsky (1967). The term is perfectly apt and useful. A bias is a systematic or predictable asymmetry, and the term is most congenial when this asymmetry applies to categories with some structural symmetry, e.g., insertions vs. deletions. The term is used this way in various areas of science and engineering, e.g., a biased estimator in statistics is one that yields a systematically low or high estimate.
Nonetheless, some evolutionary biologists don’t want you to have this useful term in your vocabulary. Some will object that “bias” should be avoided because it implies an effect on fitness, but that is just because some people think everything is about fitness and want to restrict your language to force you into their belief system. Salazar-Ciudad rejects the use of “bias” on the grounds that it implies an error or distortion. Yes, in statistics the term is used to indicate sources of distortion or sources of error from a true value, but this is a narrow 20th-century technical meaning, whereas the usage of “bias” in the English language is much older than this:
We also expect that traditionalists will dilute the concept of mutation bias as part of a cultural appropriation strategy (based on what we have seen here, here and in a recent anonymous review). That is, traditionalists will undermine the distinctive concept of mutation bias by blurring it together with chance effects, contingency, or heterogeneity, because this makes it easier for them to broaden the scientific issue and then claim that nothing is new using “we have long known” arguments, e.g., statements like “we have long known that mutation rates are not all the same” will be used to dilute the key concept, followed by “this just sounds like new words for old concepts” to undermine a claim of novelty.
The problem with this line of argument— as a critique of work highlighting the role of arrival biases— is that systematic and patterned differences in properties between classes of things are not the same thing as idiosyncratic or unpatterned heterogeneity among a set of items, and more importantly, what is novel is not the claim that mutation biases exist, but linking them with biases in evolutionary outcomes both theoretically (via a pop-gen mechanism of arrival biases) and empirically (via results showing effects of mutation bias on adaptive changes). However, the traditionalists have a lot of power, which means that they can set the terms of debate and reframe things using straw-man arguments and excluded middle arguments, e.g., “we see nothing revolutionary with X” is utterly devoid of merit but has been an effective go-to argument for traditionalists in online discussions or when talking to reporters. It’s a very easy argument to make and can be applied with to the novelty of arrival biases or other ideas. As a rhetorical device, it can be coupled very effectively with a misrepresentation of X that broadens it into something trivial, e.g., rather than saying
“we see nothing revolutionary with how this formal body of theory on arrival biases creates a structural equivalence between mutational and developmental biases that was not known to exist previously”
instead say
“we see nothing revolutionary with a theory that applies both to molecules and morphologies— we have long used such models”.
The defense of tradition often relies on fatuous arguments that broaden and trivialize new findings. Exploring them is a useful exercise to build awareness. I wish that reporters knew how to recognize this pattern of minimization.
By the way, Wikipedia gets the definition of mutation bias right. But many other sources get this wrong and say wrong things, e.g.,
- “Mutation bias. A pattern of mutation in DNA that is disproportional between the four bases, such that there is a tendency for certain bases to accumulate.” (Encyclopedia.com)
- “Mutation bias. Bias in the mutation frequencies of different codons, affecting the synonymous to nonsynonymous rate ratio. Mutation bias results in an accelerated rate of amino acid replacement in functionally less constrained regions.” [that statement is not true] (Oxford Reference)
And many sources simply do not define the term because it is not on the radar for most evolutionary biologists.
References
E. Cox and C. Yanofsky. Altered base ratios in the DNA of an Escherichia coli mutator strain. Proc. Natl. Acad. Sci. USA, 58:1895–1902, 1967.