Welcome
Welcome to the web site of the Stoltzfus research group at IBBR. Here one may find general information, links to publications, and some posts about ongoing research and other topics of interest. If you arrived at this site and haven’t found something you were looking for, please post a comment below, or send an email to Arlin. See also my ResearchGate and Google Scholar profiles.
Research
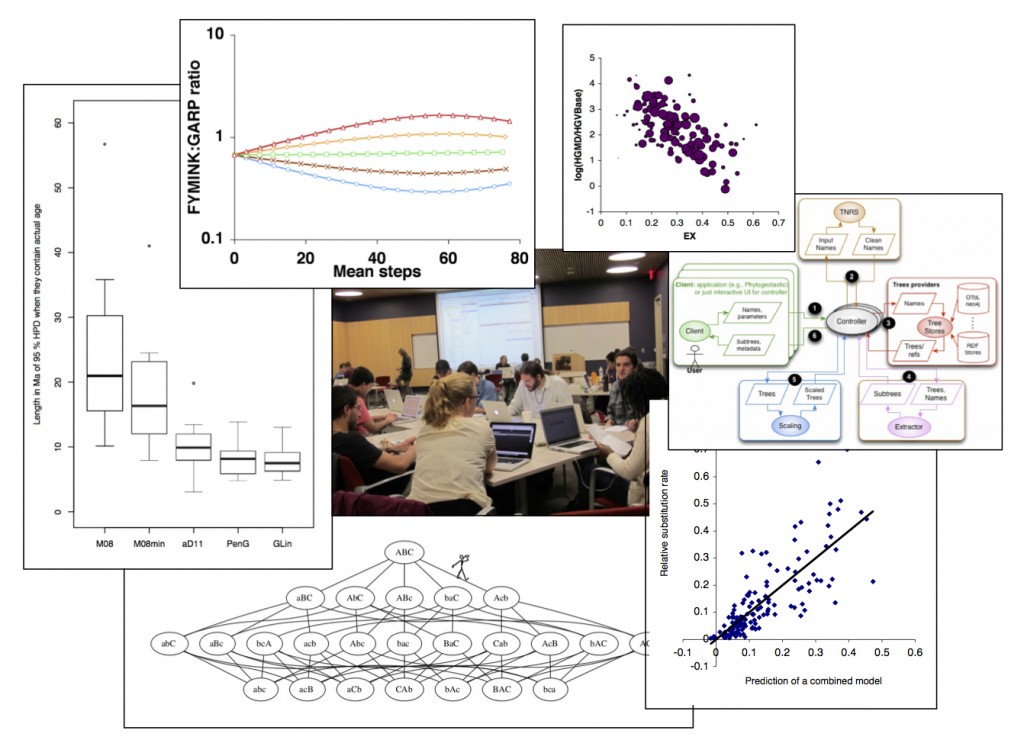 Image: A sampling of research and activities, clockwise from left
Image: A sampling of research and activities, clockwise from left
- A comparison of error in fossil-based tree calibrations using simulated data: our 2 new methods for assigning Bayesian priors from fossil intervals are on the right (Norris, et al., in review)
- a simulation of mutationally biased adaptive walks using a protein NK model: contrary to conventional wisdom, modest mutation biases (here, GC- or AT-bias) can impose a bias on the course of adaptation and cause a long-term trend (i.e., orthogenesis). See Stoltzfus, 2006.
- a validation of EX, a measure of amino acid exchangeability based on ~10K experimental exchanges: here, EX shows a strong correlation with the disease-causing propensity of amino acid changes in humans (see Yampolsky & Stoltzfus, 2005)
- high-level design of a system for on-the-fly delivery of phylogenetic knowledge by composition of web services, initially designed at the software hackathon shown in the center (see Stoltzfus, et al., 2013)
- performance of a strictly empirical (no fitted parameters) model of the relative rates of amino acid changes in human-chimp divergence (see Stoltzfus & Yampolsky, 2009)
- a drunkard’s walk through the space of scrambled genes, illustrating one of the principles of constructive neutral evolution (see Stoltzfus, 2012, or the original proposal of CNE in Stoltzfus, 1999 or Carl Zimmer’s article that fails to cite the original work or the reference in the Princeton Guide to Evolution)