Revolt of the clay: updated evidence
In a previous post called “The revolt of the clay“, I described four different ways to think about the role of variation as a process with a predictably non-random impact on the outcome of evolution. The main point was to draw attention to my favorite idea, about biases in the introduction of variation as a source of orientation or direction, and to provide a list of what (IMHO) represents the best evidence for this idea. I gave anecdotes from four categories of evidence
- mutation-biased laboratory adaptation
- mutation-biased parallel adaptation in nature
- recurrent evolution of genomic features
- miscellaneous evo-devo cases such as worm sperm
With the publication of a study by Couce, et al., 2015 (a team of researchers from Spain, France and Germany), the first category just got stronger. Couce, et al did an experiment that I’ve been trying to talk experimentalists into doing for a long time: they directly compared the spectrum of lab-evolved changes between two strains with a known difference in mutation spectrum.
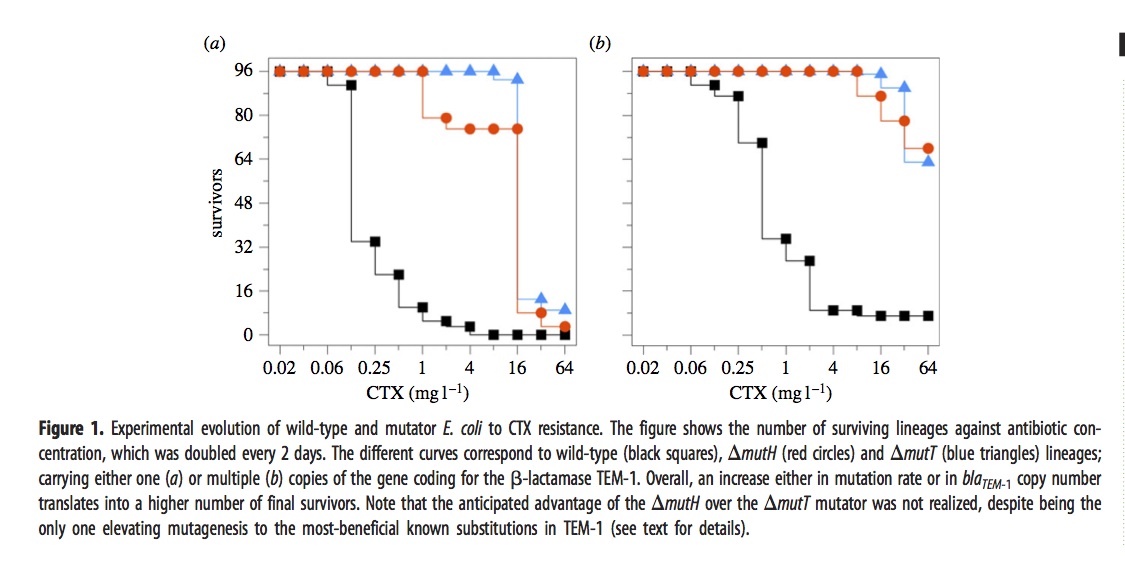
In brief, they exposed 576 different lines of E. coli to a dose of cefotaxime that started well below the MIC and doubled every 2 days for 12 iterations. The evolved lines fell into 6 groups depending on 3 choices of genetic background— wild-type, mutH, or mutT— and 2 choices of target gene copy number— just 1 copy of the TEM-1 gene, or an extra ~18 plasmid copies.
The ultimate dose of antibiotic was so extreme that most of the lines went extinct. The figure shows how many of the 96 lines (for each condition) remain viable at a given concentration.
The mutators were chosen because they have different spectra. The “red” mutator (mutH) greatly elevates the rate of G –> A transitions, and also elevates A –> G transitions. The “blue” mutator (mutT) elevates A –> C transversions. The figure above indicates that the blue mutator fared slightly better. A much stronger effect is that the lines with multiple copies of the TEM-1 gene did better. Very few single-copy strains survived the highest doses of antibiotic.
The authors then did some phenotypic characterization that led them to suspect that it was not just the TEM-1 gene, but other genes that were important in adaptation. In particular, they suspected the gene for PBP3, which is a direct target of cefotaxime.
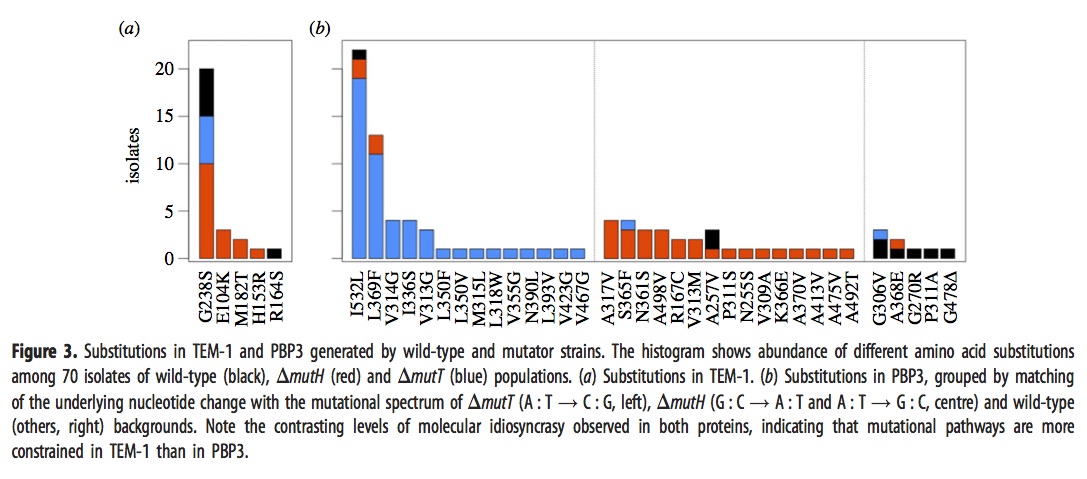
They sequenced the genes for TEM-1 and PBP3 from a large sample of the surviving strains above, with results shown in this figure:
The left panel, i.e., figure 3a, shows that evolved strains of mutH (red), mutT (blue), and wild-type (black) sometimes have one of the well-known mutants in the TEM-1 gene encoding beta-lactamase. The predominance of red means that more mutH strains evolved the familiar TEM-1 mutations, which makes sense because these mutations happen to be the G–>A and A–>G transitions favored by mutH.
Panel 3b, which refers to mutants in the gene encoding PBP3, is a bit complicated. As before, the histogram bars are colored based on the background in which the particular mutation listed on the x axis is fixed. However, rather than showing all the lines together in one histogram, they are arranged into 3 different histograms depending on whether the mutational pathway matches the pathways preferred by mutT (left sub-panel), by mutH (center sub-panel), or neither (right sub-panel). For instance, blue appears in 16 columns, so there are 16 different mutant PBP3 types that evolved in mutT backgrounds, and 14 of them are in the left sub-panel indicating mutT-favored mutational pathways. Red appears in 18 columns, 15 corresponding to mutH-favored pathways.
Importantly, the density in the columns doesn’t overlap much: nearly all of the blueness is in the left histogram (~50/52), and nearly all the redness is in the center histogram (~26/31). This means that evolved mutT strains overwhelmingly carried mutations favored by the mutT mutators, and likewise for mutH strains and mutH mutators. If adaptation were unaffected by tendencies of mutation, as the architects of the Modern Synthesis believed, then the colors would be randomly scrambled between the 3 histograms. Because it is not, we know that the regime of mutation imposes a predictable bias on the spectrum of adaptive changes.
The main weakness of the paper, perhaps, is that they did not do reconstructions to show causative mutations. Instead they make plausibility arguments based on having seen some of these in other studies of laboratory or clinical resistance. I think the paper is pretty strong in spite of this. Recurrence of a mutation in independent lines is a very strong sign that it is causative, whereas singletons might be hitch-hikers with no causative significance. Yet if we were to eliminate the 17 non-recurring mutants, we would still have a striking contrast. Of ~43 mutT mutants with recurrently fixed mutations, ~41 are for mutT-favored pathways; of the ~23 mutH mutants with recurrently fixed mutations, ~18 are for mutH-favored pathways.
Unfortunately, due to the language and framing of this paper, casual readers might not recognize that it is actually about mutation-biased adaptation. The term “mutation bias” only occurs once, in the claim that “Mutational biases forcibly impose a high level of divergence at the molecular level.” I’m not sure what that means. What I would describe as “evolution biased by mutation” is described by Couce, et al as “limits to natural selection imposed by genetic constraints”. Sometimes the language of my fellow evolutionists seems like a strange and foreign language to me. But I digress.
To summarize, this work clearly establishes that 2 different mutator strains adapting to increasing antibiotic concentrations exhibit strikingly different spectra of evolved changes that match their mutation spectra, consistent with the a priori expectation of mutation-biased adaptation.