The revolt of the clay: an initial progress report
A “chance” encounter
Earlier this month I was contacted by a reporter writing a piece on the role of chance in evolution. I responded that I didn’t work on that topic, but if he was interested in predictable non-randomness due to biases in variation, then I would be happy to talk. We had a nice chat last Friday.
I’m only working on the role of “chance” in the sense that, in our field, referring to “chance” is a placemarker for the demise of an approach based implicitly on deterministic thinking— evolution proceeds to equilibrium, and everything turns out for the best, driven by selection. This justifies the classic view that “the ultimate source of explanation in biology is the principle of natural selection” (Ayala, 1970). Bruce Levin and colleagues mock this idea hilariously in the following passage from an actual research paper:
To be sure, the ascent and fixation of the earlier-occurring rather than the best-adapted genotypes due to this bottleneck-mutation rate mechanism is a non-equilibrium result. On Equilibrium Day deterministic processes will prevail and the best genotypes will inherit the earth (Levin, Perrot & Walker, 2000)
Wait, I’m still laughing. OK. Much evolutionary research still takes place within an adaptationist research program whose goal is to explain “phenotype existence” by invoking selection (Reeve & Sherman, 1993). Framing the goal of evolutionary research as providing raisons d’être is IMHO a mistake. Instead of rejecting raisons d’être, leading thinkers like Ernst Mayr made the pursuit of natural purposes an official policy, presenting an allegedly materialistic version of selection-derived “function” to stand as an alternative to purposive creation. The problem is that the theory underlying this version of teleology, if it works at all, only works on Equilibrium Day.
An alternative is to approach evolution as change, rather than as the reason things exist. But instead of tossing out this old framework, we are just patching it up with “chance”, “constraints”, and “contingency”— i.e., a bunch of excuses, not legitimate causal processes. I suppose it represents progress that, instead of an inadequate default view of evolution, the past 30 years have brought us something different— an inadequate default view supplemented with concepts that betoken its inadequacy. In short, I’m not really working on chance, but in my field, anyone who is not working on selection is said to be working on the role of chance.
 After my little screed about “chance”, the discussion turned to my favorite hobby horse, the idea that biases in variation play a large and unappreciated role in evolution. Eventually we got to the topic of what was the best evidence for the idea. I can’t remember what I said on the phone, but I thought this question deserved a more dedicated effort, so I gave it some more thought and wrote a followup email that listed 5 kinds of evidence for the specific idea that Lev Yampolsky and I proposed (Yampolsky and Stoltzfus, 2001).
After my little screed about “chance”, the discussion turned to my favorite hobby horse, the idea that biases in variation play a large and unappreciated role in evolution. Eventually we got to the topic of what was the best evidence for the idea. I can’t remember what I said on the phone, but I thought this question deserved a more dedicated effort, so I gave it some more thought and wrote a followup email that listed 5 kinds of evidence for the specific idea that Lev Yampolsky and I proposed (Yampolsky and Stoltzfus, 2001).
And then, having spent time on that, I was inspired to write this post.
The revolt of the clay
To start, let’s compare and contrast ideas about the role of variation in evolution. The canonical neo-Darwinian view, as described in Gould’s giant book (pp 137 to 146), is that variation is abundant, infinitesimal, and random [1]. When these conditions are met, “selection” can be construed as an agent that supplies initiative, creativity and direction to evolution, while variation supplies none of these things, merely providing passive raw materials. Selection is the potter, and variation is the clay. Selection shapes adaptations out of the raw materials of variation.

Poulton was poking fun at non-Darwinian views with his phrase “the revolt of the clay,” whereas for me, the Darwinian metaphor of Selection as Craftsman or Artist (see Gould, 1983) is such a deserving target of parody that I’d like to keep “revolt of the clay” alive. Unmodified image from AR Nature Gal, https://www.flickr.com/photos/naturegal/3364160454/, some rights reserved
Evolution doesn’t have to work this way. And if these Darwinian restrictions on the role of variation are not met, selection can’t be invoked creatively in the same way. If variation isn’t like the clay, selection isn’t like the potter who shapes it.
A century ago, Poulton referred to non-Darwinian views of variation as “the revolt of the clay against the power of the potter” (image). Today, advocates of non-Darwinian views of variation often invoke de Vries’s emphasis on “the arrival of the fittest”. This new revolt of the clay includes several ideas:
- The G matrix is profoundly important as a predictor of quantitative trait evolution. Classical quantitative genetics comes from Fisher’s infinitesimal view, which doesn’t sound very radical, and isn’t. In the classic model, variation in a continuous trait (e.g., height) is normally distributed, with a mean effect of zero, thus variation cannot be a source of direction. However, the multivariate generalization of quantitative genetics (Lande and Arnold, 1983), which addresses multiple characters simultaneously, introduces something a bit subversive. Although the mean effect of variation in every dimension is still zero, the variances and covariances in the multivariate “G matrix” of variation differ. This makes variation something more than passive clay: as Steppan, et al say, “Together with natural selection (the adaptive landscape) it determines the direction and rate of evolution.” For instance, there is an axis in the multi-dimensional space of characters with the most variance, Gmax, and we expect the maximum of evolutionary change to occur along this axis— this effect of “evolution along genetic lines of least resistance” is well documented (Schluter, 1996, McGuigan, et al., 2005).
- Mutation is “directed” (Cairnsian, adaptive). Here the idea is that the process of mutation is jiggered to provide situation-appropriate mutations. For instance, the view of James Shapiro is that mutations should be seen, not as accidents, but as “natural genetic engineering”. The default Darwinian view is very strongly externalist— organisms are shaped by external conditions, and internal forces are viewed with suspicion as an appeal to vitalism. However, in Darwinian evolution by natural selection, externalities are internalized only by selection, while variation is asserted to be “random” relative to externalities. By contrast, the “directed” view is that mutation systems also internalize information on external conditions, an effect that is well documented in some cases (e.g., yeast mating-type switching or the crispr-cas system). For a nice introduction, see the recent article in Quanta (image).
 As a position on evolution, this view holds that (1) “mutation is random” is a core doctrine of Darwinism that (2) has been refuted empirically, therefore (3) we must reject Darwinism and rethink evolution. This quixotic line of argument has led to more heat than light. Today’s DiNOs (Darwinians in Name Only) will never give up the label “Darwinian”, and they would rather redefine “random” to include directed mutation than to give up a cherished doctrine (see Merlin, 2010). Directed mutation people, I’m talking to you now. You can’t win this argument. You can’t convince mainstream evolutionary biologists to re-think evolution on the grounds of having found something new about mutation (or ecology or physiology or any other not-evolution field). This is not just because they are committed to believing that mutation is unimportant (which is what “mutation is random” really means), but because this is the way that science works. You need to re-think evolution (and you need to do a way better job than Shapiro). You need to figure out the theoretical consequences of directed mutation for evolution, then do comparative evolutionary research showing that those consequences have been important. Then you will have an argument to make in the field of evolutionary biology.
As a position on evolution, this view holds that (1) “mutation is random” is a core doctrine of Darwinism that (2) has been refuted empirically, therefore (3) we must reject Darwinism and rethink evolution. This quixotic line of argument has led to more heat than light. Today’s DiNOs (Darwinians in Name Only) will never give up the label “Darwinian”, and they would rather redefine “random” to include directed mutation than to give up a cherished doctrine (see Merlin, 2010). Directed mutation people, I’m talking to you now. You can’t win this argument. You can’t convince mainstream evolutionary biologists to re-think evolution on the grounds of having found something new about mutation (or ecology or physiology or any other not-evolution field). This is not just because they are committed to believing that mutation is unimportant (which is what “mutation is random” really means), but because this is the way that science works. You need to re-think evolution (and you need to do a way better job than Shapiro). You need to figure out the theoretical consequences of directed mutation for evolution, then do comparative evolutionary research showing that those consequences have been important. Then you will have an argument to make in the field of evolutionary biology. - Variation is “facilitated”; natural developmental-genetic systems are “evolvable”. This is how evo-devo people talk about variation now, replacing their older language of “constraints” [3]. To understand this idea can be very difficult. I’ve been trying for years and I am not sure that I have succeeded. The position of Kirschner and Gerhardt differs from Shapiro’s in that they insist that variation remains “random” at the “genetic” level, while the developmental “response” to genetic variation enhances the production of appropriate phenotypic variations. In Andreas Wagner’s latest book (figure), the focus is on “natural principles that accelerate life’s ability to innovate” (last page of the intro).
 Other authors take slightly different positions. In this view, the facilitated nature of variation, the “evolvability” of systems, is a kind of internal organizational principle that has been overlooked: the needed reform of evolutionary theory consists in integrating this new principle. Thus, what distinguishes the evo-devo position from mere quantitative genetics (above) is the view that evolution has been facilitated or speeded up or improved because of the way that developmental-genetic systems tend to produce more of the right kinds of variations. Whereas directed mutation proponents generally are adaptationists who believe that directed mutation is an adaptation for evolving, Kirschner and Gerhard refuse to take a position on whether facilitation is an adaptation. To use the language of philosophers, the explanandum (the thing-to-be-explained) is that organisms are so awesomely “evolvable” or “innovable” (A. Wagner), but the evo-devoists are not seeking the kind of adaptationist “why” that would explain evolvability as an adaptation: instead they want a kind of mechano-structuralist explanation, an explanation in terms of developmental principles.
Other authors take slightly different positions. In this view, the facilitated nature of variation, the “evolvability” of systems, is a kind of internal organizational principle that has been overlooked: the needed reform of evolutionary theory consists in integrating this new principle. Thus, what distinguishes the evo-devo position from mere quantitative genetics (above) is the view that evolution has been facilitated or speeded up or improved because of the way that developmental-genetic systems tend to produce more of the right kinds of variations. Whereas directed mutation proponents generally are adaptationists who believe that directed mutation is an adaptation for evolving, Kirschner and Gerhard refuse to take a position on whether facilitation is an adaptation. To use the language of philosophers, the explanandum (the thing-to-be-explained) is that organisms are so awesomely “evolvable” or “innovable” (A. Wagner), but the evo-devoists are not seeking the kind of adaptationist “why” that would explain evolvability as an adaptation: instead they want a kind of mechano-structuralist explanation, an explanation in terms of developmental principles. - Bias in the introduction of variation is an important orienting factor in evolution. Imagine a world of discrete types in which the chance of getting from X to Y is just the rate of mutation from X to Y, multiplied by the chance a mutant Y will take over a population of Xs. A mutational bias that makes mutation B times more likely than to go to Y1 than Y2 has exactly the same impact on the course of evolution as a fitness effect that makes it more likely for Y1 to be fixed than Y2. This way of thinking actually corresponds to a special case of “mutation-limited” population genetics. It isn’t very realistic, but it helps us to understand a more general idea first presented by Yampolsky and Stoltzfus (2001), in which tendencies of evolution are understood to be a dual function of biases in the introduction of variants (by mutation and altered development) and biases in the reproduction of variants (i.e., selection). The figure below shows the simplest possible population model of this effect, in which evolution can climb to the left (from ab to Ab) or climb to the right (from ab to aB), where the right peak is higher but the rate of mutation to the left is higher by a factor of B. The main point of the figure below is that as B increases, the system tends to evolve to the mutationally favored peak, even though selection is operating and the population is adapting by going to a more fit genotype.
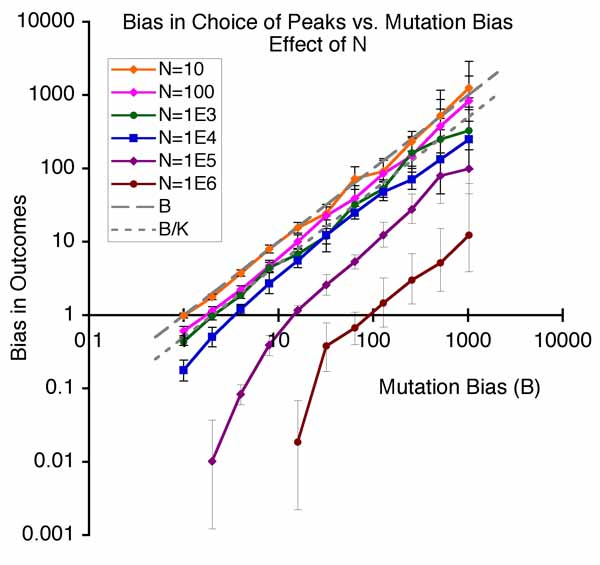 Because evolutionary biologists just don’t tend to think this way, this basic principle was novel when proposed in 2001, and still isn’t explained in textbooks. The focus here is not on identifying a class of mutations that internalize external information, or on organizational principles that make evolution better. Instead, as with quantitative genetics, there is no assumption that patterns of variation make evolution better: they are simply biases or non-uniformities— whether stupid ones or smart ones— with a predictable impact on the course of evolution. Unlike quantitative genetics, the focus is on biases along an axis, not simply differences in total amount of variation for a character. The explanandum is not why evolution happened so fast (relative to some hypothetical world where it is slower), but why evolution has the tendencies it has, i.e., why it tends to take one course more than another. This involves understanding both the developmental-genetic causes of biases, and their population-genetic consequences.
Because evolutionary biologists just don’t tend to think this way, this basic principle was novel when proposed in 2001, and still isn’t explained in textbooks. The focus here is not on identifying a class of mutations that internalize external information, or on organizational principles that make evolution better. Instead, as with quantitative genetics, there is no assumption that patterns of variation make evolution better: they are simply biases or non-uniformities— whether stupid ones or smart ones— with a predictable impact on the course of evolution. Unlike quantitative genetics, the focus is on biases along an axis, not simply differences in total amount of variation for a character. The explanandum is not why evolution happened so fast (relative to some hypothetical world where it is slower), but why evolution has the tendencies it has, i.e., why it tends to take one course more than another. This involves understanding both the developmental-genetic causes of biases, and their population-genetic consequences.
Others might describe these views differently, and each has a different status in evolutionary thinking today. The quantitative genetics stuff is covered in textbooks. Multiple books and reviews have been written about evolvability, and directed variation has been debated extensively in journals. There is even a wikipedia page on “evolutionary capacitance“, a variation-related idea that isn’t on the list above. I would not expect anyone to be aware of my pet idea above, which is referenced only in a Wallace Arthur’s Biased Embryos and Evolution.[4] For most evolutionary biologists, #1 is established knowledge, #2 is (rather unfairly) ignored as fringe science, #3 is accepted as the latest news from evo-devo— albeit with an unclear significance—, and #4 isn’t on the radar.
Evidence
On what basis could one construct an evidentiary case for the importance of biases in variation as a cause of orientation in evolution?
The practical problem, when trying to make a convincing case, is that we (scientists) don’t want to rearrange our mental furniture every month to adjust to important new findings, so we tend to resist anything that would do so, including by labeling it “unimportant”. If I report some tendency of evolution E and show that E is due to mutational bias, the likely response from a mainstream reader is to conclude that E is unimportant, which not only protects the reader from having to rearrange his mental furniture, but also relieves him of any need to evaluate the rigor of a claim that, after all, isn’t important.[5]
For reasons that I’m not going to explain, mutation-biased neutral evolution does not require us to rearrange our mental furniture. No one cares if pseudogenes and spacers show effects of mutation bias. So, I’m not going to bring forward any of the voluminous (albeit largely circumstantial) evidence for compositional biases in genomes caused by nucleotide mutation biases.
Instead I’m going to focus on cases where evolutionary change is clearly adaptive, or where the hypothesis of a mutational cause stands as an alternative to a selective explanation.
Mutation-biased adaptation in the lab
Various experimental evolution studies show the effect of biases in mutation, but almost none of them report it because this issue is not on the radar. The sole exception, to my knowledge, is the study of phage parallel adaptation by Rokyta, et al 2005. The most frequently recurring genotype was not the one with the highest selection coefficient, but was favored by a considerable mutation bias. A model that ignored mutation was rejected in favor of a model that includes transition-transversion bias.
In all other cases, mutational effects are not noted and one has to analyze the data oneself. 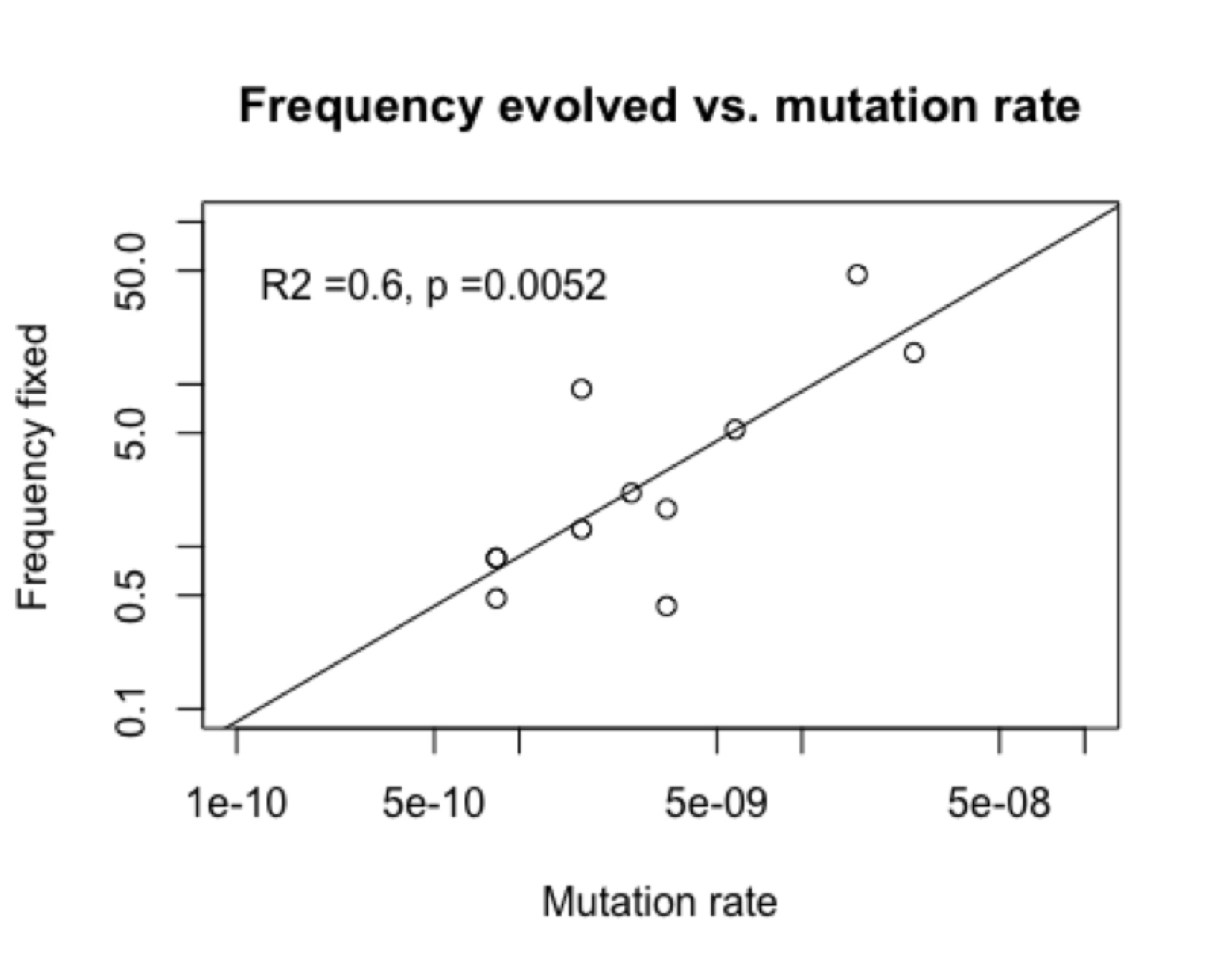
The figures below show an example from a published study of Rif-resistance evolution in Pseudomonas aeruginosa. The chance of finding a particular genotype among the adapted strains (“frequency fixed”) is significantly correlated (figure, right) with the rate of mutation to that genotype reported by the same authors in the same study. 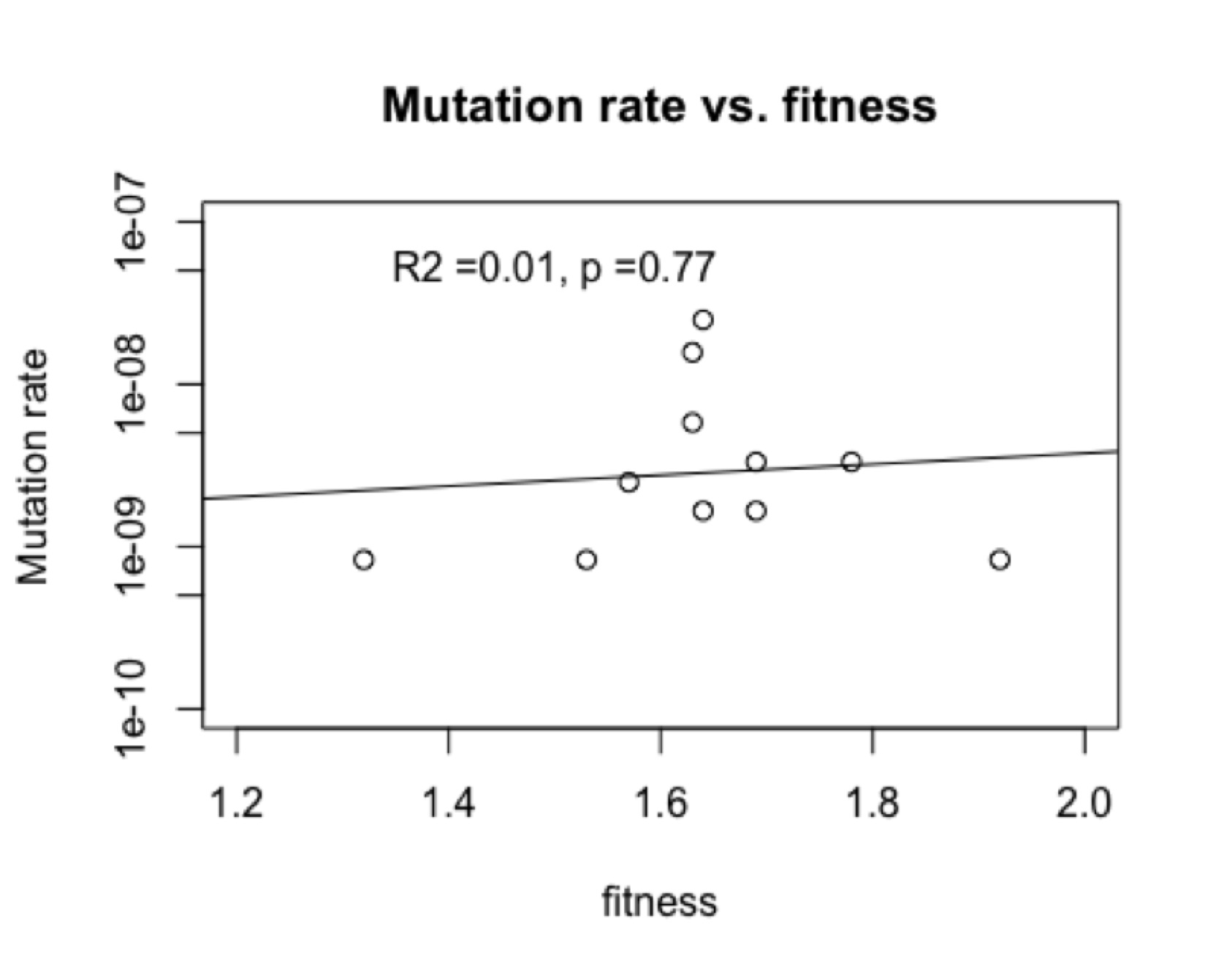
This is not due to “directed mutations” because in fact (figure, left) there is no correlation of mutation rate and fitness. I showed this to the author and asked if he wanted to explore this revolutionary new phenomenon, and he replied that he wasn’t working on this system anymore. Apparently, documenting a new kind of evolutionary cause is not a worthwhile use of time if you are hot on the trail of something really novel and profound like . . . epistasis.
Mutation-biased parallel adaptation in nature
Last year I combed through reported cases of parallel adaptation in nature, finding cases in which a phenotype-first approach is used to identify exactly parallel amino acid replacements. Some of these are famous cases like parallel lysozyme changes in foregut-fermenting organisms, altitude-adapted hemoglobins in birds, and spectral tuning of photoreceptors. There are enough cases now that one can actually do statistics— my latest compilation implicated 94 replacements (in 16 different proteins) that occurred on average 4 times each (347 separate evolutionary events). I simply asked whether the transition:transversion ratio among these replacements is higher than the ~0.4 ratio that one expects if there is no mutation bias. In fact, the ratio is about 1, i.e., 2.5-fold higher than expected without mutation bias. There isn’t a publication yet, but there is a short video based on my presentation at SMBE last June (this combines natural and experimental cases: near the end, I note that the conclusion holds when experimental cases are excluded).
Recurrent evolution of miscellaneous genomic features
A variety of cases of recurrent evolution of genomic features are reviewed and analyzed by Maseo, et al. (the figure at right is from their paper). I won’t go into detail on individual cases because the paper aims more to draw attention to a new area of inquiry (by defining and classifying patterns) than to establish particular causal mechanisms in any specific case. However, in many cases the mutational explanations seem plausible, e.g., the proposal of Roy and Penny that extensive genome-wide loss of introns might be a mutational effect due to runaway retroposition (by which intronless cDNAs can get introduced back into the genome, resulting in intron loss).
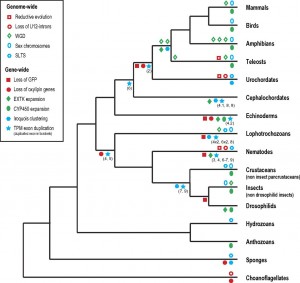
An example not discussed by Maseo, et al is a recurrent pattern in the evolution of mitochondrial genomes: a remarkably biased flow (over 100s of millions of years) of genes from the mitochondrion to the nucleus, and not in the reverse direction. This could be affected by many different factors, one of which is asymmetry in rates of DNA transfer of gene-sized fragments. In a study with yeast, Thorsness and Fox (1990) found the rate of mito-to-nuc transfer was at least 105-fold higher than nuc-to-mito transfer. Berg and Kurland (2000) used these rates in an evolutionary model for the fate of mitochondrial genes. Where M and m are active and inactive mito, and likewise N and n are active and inactive nuclear copies, the model is simply:
where the c’s are rates of mutational transfer and establishment, and the u’s are rates of inactivation. One of the conclusions of the authors is that the bias in transfer rates is important in establishing directionality. [6]
Preferences for regulatory or structural changes
The issue of an evolutionary preference for regulatory vs structural changes— whether a preference exists, and if so, why— has been debated for decades. The issue has received renewed attention in evo-devo in the past decade. Streisfeld and Rausher (2011) attempt to tackle the causes of preferences quantitatively, introducing a previously missing framework to evaluate mutational and selective causes of biases between types of changes. The missing framework is actually the origin-fixation framework that has existed since 1969 but is rarely recognized as a coherent theory, and the method of apportioning evolutionary preferences into biases in fixation and biases in mutation is the same as in Yampolsky & Stoltzfus. They examine evidence from floral evolution and find evidence for both mutational and selective sources of bias.
Sexy worm stuff
There are also studies on worms in which the authors argue that developmental effects on variation cause a bias toward certain types of evolutionary changes (what makes this work sexy is only that so much of the developmental work on worms involves sex-determination and the development of gonads and secondary sexual characters). The best one I have seen is “A Bias Caused by Ectopic Development Produces Sexually Dimorphic Sperm in Nematodes” (Baldi, Viviano & Ellis, 2011). There have been multiple cases in which Caenorhabditis species evolved androdiecy, with hermophrodites that produce sperm as late-stage larvae, then go on to act like females, producing eggs as adults. In every case, the hermaphrodite sperm are substantially smaller than male sperm, an effect that has been attributed to selection favoring male over hermaphrodite sperm. Previously, Baldi, et al reported that RNAi directed against a sex-determining gene (tra-2) caused some females of non-hermaphroditic species to become pseudo-hermaphrodites, making sperm as juveniles before becoming female adults (i.e., just like natural hermaphrodites). Here they report that, in the 3 cases tested, pseudo-hermaphrodites make smaller sperm than the corresponding males (figure: the top panel shows sperm size for C elegans, which is naturally hermaphroditic; the other three are experimentally induced).
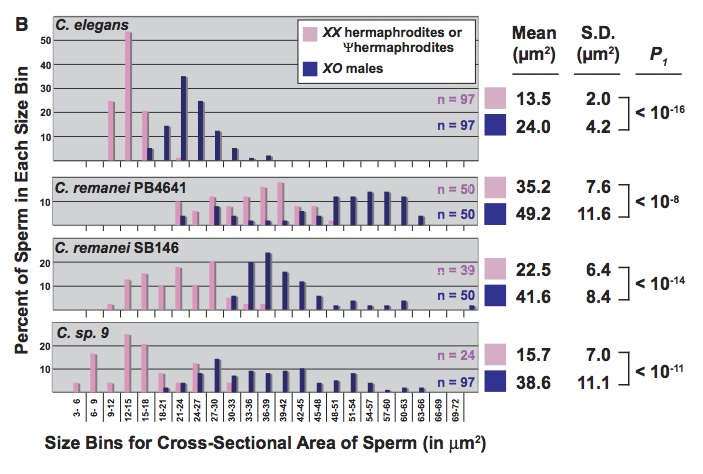 This can’t be due to selection because these are novel phenotypes in species that do not have hermaphrodites naturally. That is, there appears to be a developmental bias toward smaller sperm in novel hermaphrodites, and the authors suggest that this bias has contributed to the observed evolutionary bias.
This can’t be due to selection because these are novel phenotypes in species that do not have hermaphrodites naturally. That is, there appears to be a developmental bias toward smaller sperm in novel hermaphrodites, and the authors suggest that this bias has contributed to the observed evolutionary bias.
In “Bias and Evolution of the Mutationally Accessible Phenotypic Space in a Developmental System”, Braendle, Baer, and Félix (2010) report the results of a mutation-accumulation study accompanied by analysis of phenotypic effects and natural variation. Their deliberate aim is to characterize the spectrum of mutational effects, focusing on vulval development (the specialty of the Félix lab). After presenting these results, they go on to address how such biases might have affected evolution. Unfortunately for my pet idea, when the authors conclude that “evolutionary trends in the vulval system concern the phenotypic characters that are most easily affected by mutation”, they are really talking about differences in the amount of variation that can be understood under the quantitative genetics model or “evolvability” view.
Conclusions
This is a progress report on what I think is a promising and potentially transformative idea, but there is apparently only 1 person in the world working on it, so progress is . . . s . . . l . . . o . . . w. To put things in the immortal words of Gil Scott Heron, the Revolution Will Not Be Televised.
None of the cases discussed above are examples of quantitative genetics, or directed mutation, or “evolvability”. Instead they are (putative) examples of evolution biased by the introduction of variation.
Each of the above examples has strengths and weaknesses. The laboratory evolution cases clearly provide strong and direct evidence that the things that turn up in adaptation tend to be mutationally likely, and this is not because of a correlation with fitness, but because their mutational likelihood alone is a non-selective evolutionary advantage, a direct kinetic bias on the process of adaptation. But these are laboratory cases, and ideally one wants something from nature. I’m excited about the cases of natural parallel evolution, although there are a lot of ambiguities involved in the analysis of these data.
In the remaining cases, I think the connection between the evidence and a particular mechanistic cause of evolutionary bias is not well established. The case of sperm-size bias is interesting, however, because the variational hypothesis is an alternative to a selective hypothesis already in the literature.
By the way, I don’t think there is anything special about worms. It just happens to be the case that a few people in the worm development field (Marie-Anne Félix, Christian Braendle, Eric Haag) noticed Yampolsky & Stoltzfus (2001) and started citing it. I identify these cases via citations. This raises the question of whether I might be missing all sorts of other cases in which authors are not citing these works, but are reporting biases in variation and relating them to evolutionary biases. I don’t know the answer to that question, but I suspect the answer is negative. The reason for this is that I periodically search the web with various phrases like “mutation-biased adaptation” or “mutation bias and evolution” in order to turn up examples, and I usually don’t get anything I don’t know about already.
Footnotes
[1] Gould actually uses the words “copious”, “undirected” and “small in effect”, on the way to making the same argument about creativity. Gould argues that the essence of Darwinism is that “selection creates the fit”. I don’t believe in essences, and neither does Gould, but this is a very helpful way of characterizing Darwinism if one wants to understand its history in the face of alternative views. Contemporary advocates of Darwinism do not openly defend the creativity doctrine, but it played a huge role in shaping Darwinism.
[2] Note a few things. G_max is an axis, not a direction: a line, not an arrow. Also, it is a measure of standing variation, not a mutation spectrum, though one can consider models that include an M matrix of new variation. Finally, the reason that we expect the correlation between G_max and evolutionary divergence is that we have abandoned the idealistic Panglossian view in which evolution would produce the optimal phenotype even if that required moving along an axis that has very little variation. Instead, as Steppan et al say, the rate and direction of evolution are determined jointly by selection and variation. That is why this model goes into the “revolt of the clay” category, even though the field of quantitative genetics is clearly the offspring of neo-Darwinian orthodoxy. Sometimes children rebel. In general, evolutionary biologists have responded to the breakdown of the original neo-Darwinian view by re-defining it to mean whatever is left of the original neo-Darwinian view, though this is increasingly indistinguishable from mutationism.
[3] Note that there is more to evo-devo than the non-canonical position on the role of variation that is my focus here. Evo-devo also represents a position about the kinds of causal explanations that are important.
[4] The 3 papers that express my pet idea have been cited ~100 times total. That’s well above average, but it doesn’t amount to widespread recognition. When this work is cited in the evo-devo literature, it typically is not distinguished from other ideas combining development, variation and evolution. Some citations convey misconceptions, e.g., Martincorena and Luscombe write “For decades, the question of how selection affects mutation rates has attracted considerable attention from evolutionary biologists”, citing over 20 works including ours.
[5] I realized this after a particular seminar in which I had shown that patterns of amino acid replacement in human-chimp divergence were profoundly affected by mutation biases in a way that was quantitatively predictable (see Stoltzfus & Yampolsky 2009). A junior professor stood up after the talk, and— with an air of breaking through the fog of my talk to reflect on the true nature of my findings— just kept repeating words to the effect that “Yes, it happens, but it isn’t important! It isn’t important!”.
[6] At the risk of confusing things, I have to say something potentially confusing about this. There are actually 2 kinds of biased mutational effects in the Berg-Kurland model, but only one of them corresponds to the Yampolsky-Stoltzfus effect. The Yampolsky-Stoltzfus effect refers to an evolutionary bias among alleles destined for fixation, whereas one also can consider a mutational bias leading to differential loss of alleles. In classical population genetics, mutation is often treated as a mass-action deleterious pressure, as in the mutation-selection balance equation f = u/s (deleterious mutation drives frequency up, selection drives it down). Much of Michael Lynch’s reasoning about the evolution of genome organization depends on the differential pressure of deleterious mutation on excess DNA— longer alleles have a built-in cost due to additional mutations. In the simplest version of the Berg-Kurland model, the inactivation rate is simply a form of this: an extra copy can get degraded through mutation (in the more complex version, this process can be speeded by selection, e.g., for a shorter genome). Berg and Kurland believe that inactivation is biased to favor transfer to the nucleus, because the mitochondrial mutation rate is higher (i.e., genes get knocked down by mutation faster). Because my comments here are focused on the Yampolsky-Stoltzfus effect, I am emphasizing only the bias in rates of mutational transfer from one compartmental genome to another, not the bias in rates of mutational inactivation that is also a mutational effect. But more generally, Berg and Kurland conclude that both biases are important.
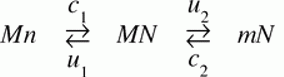
Comments